Scientists Develop Machine-learning-backed Model of Protein Behavior Linked to Cancer-causing Mutations
 A multi-institutional team of scientists have developed a highly detailed, machine learning-backed multiscale model revealing the importance of lipids to the signaling dynamics of RAS, a family of proteins whose mutations are linked to numerous cancers.
A multi-institutional team of scientists have developed a highly detailed, machine learning-backed multiscale model revealing the importance of lipids to the signaling dynamics of RAS, a family of proteins whose mutations are linked to numerous cancers.
Published by the Proceedings of the National Academy of Sciences, the paper from researchers led by the Lawrence Livermore National Laboratory details the methodology behind the Multiscale Machine-Learned Modeling Infrastructure (MuMMI), which simulates the behavior of RAS proteins on a realistic cell membrane, their interactions with each other and with lipids— organic compounds that help make up cell membranes — and the activation of signaling through the RAS interaction with RAF proteins, on a macro and molecular level.
It also discusses the team’s findings from using the framework to model how RAS binds to other proteins and how different kinds of lipids dictate how RAS collects and positions itself on the cell membrane. Evaluating tens of thousands of simulations, the team captured all previous protein interactions and many more RAS interfaces. The data indicates that lipids —rather than protein interfaces — govern both RAS orientation and accumulation of RAS proteins.
Normally, RAS receives and follows signals to switch between active and inactive states, but as the proteins move along the cell membrane — like balls of string tumbling along a fluid ground — they combine with other proteins and can activate signaling behavior. Mutated RAS proteins can become stuck in an uncontrollable, “always on” growth state. This state is indicated in the formation of about 30 percent of all cancers, particularly pancreatic, lung and colorectal cancers.
Researchers said the MuMMI framework represents a “fundamentally new technology in computational biology” and could be used to inform new experiments and improve scientists’ basic understanding of RAS protein binding. Previous scientific literature has proposed numerous orientations for how RAS comes together, with a major hypothesis being that there is some preordering of RAS proteins on the membrane prior to downstream signaling.
“We always knew lipids were important; you need some of them, otherwise you don’t have this behavior. But after that, scientists didn’t know what was important about them,” said LLNL computer scientist and first author Helgi Ingolfsson. “This work is showing us that lipids are a key player. By modulating the lipids and different lipid environments, RAS changes its orientation, and you can actually change the signaling [between ‘grow’ and ‘not grow’] by changing the lipids underneath. Now we have an enormous sample of simulations, and we can see how RAS interacts in all our simulations at different angles. The message is that yes, they come together, but they come together in all kinds of different orientations.”
The paper is part of an ongoing pilot project of the Joint Design of Advanced Computing Solutions for Cancer (JDACS4C) collaboration between the Department of Energy, the National Cancer Institute (NCI) and other organizations, it includes co-authors at the NCI’s Frederick National Laboratory for Cancer Research (FNLCR) who are applying some of insights gained from the model in lab experiments.
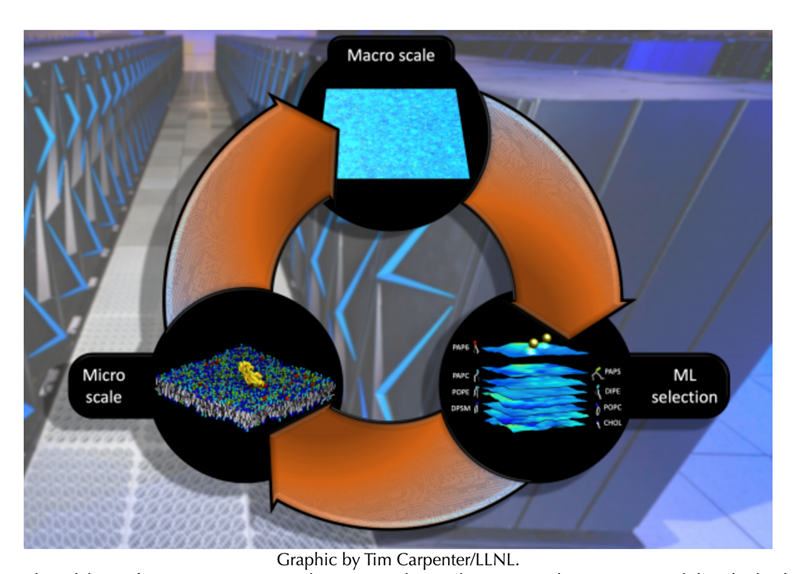
MuMMI’s ability to provide insights at two different temporal and spatial scales allowed the team to examine thousands of different RAS-lipid compositions and observe distinct interaction patterns and numerous RAS orientations. Starting with a broad macroscale model, a machine learning algorithm automatically selected lipid “patches” it deemed interesting enough to examine more closely with the micromodel simulations.
The team simulated a one micron-by-one micron patch on LLNL’s Sierra supercomputer and observed how hundreds of different RAS proteins interacted with eight kinds of lipids. They created more than 100,000 smaller molecular dynamic simulations from machine learning-selected “interesting” snapshots of the larger macro model simulation, enabling them to determine the probabilities of RAS binding to other proteins with a given orientation on a cell membrane.
Scientists at FNLCR performed the microscopy, biophysical, biochemical and structural biology experiments needed to parameterize the simulations. Combined with experimental results, the work demonstrates the strong link between lipids and RAS orientation and binding probability. Researchers found only specific RAS orientations could bind with other proteins to induce signaling behavior and that binding probability is lipid-dependent — knowing only lipid compositions, scientists could predict the orientation of RAS on the membrane with high fidelity.
“Scientists know that RAS has to create the signal, and they know RAS has to meet another RAS, but they don’t know why, and they don’t know necessarily how at an atomistic level,” said co-author and LLNL Biochemical and Biophysical Systems Group Leader Felice Lightstone. “The insights here confirmed experimental results that are always controversial when you don’t have really precise measurements. For the RAS signaling pathway to continue, you need to bind to a RAF, and certain orientations make it impossible to bind and continue the signal.”
Traditionally, scientists simulate only a small, fixed number of proteins and one lipid composition, Ingolfsson explained, and need to know which lipids are important to model beforehand. With MuMMI, researchers can simulate thousands of different cell compositions derived from the macro model, allowing scientists to answer questions about RAS-lipid interactions that could only be possible with a multiscale simulation, researchers said. In the future, Ingolfsson said, scientists won’t do one simulation at a time, but an entire ensemble of simulations, selecting the most interesting areas with machine learning algorithms.
“We’re demonstrating that the old way of doing things is starting to be outdated,” Ingolfsson said. “At Livermore, we have enormous computing power, we have a lot of people working on this and we can show what can be possible.”
Researchers said the insights from MuMMI also will be useful for experimentalists, who are generally limited to testing 1 or 2 lipid types due to cost or complexity. Experimentalists typically use regular cells, which include everything, or create simple model systems that don’t capture all the necessary data, Lightstone said. With the multiscale model, the team can generate new hypotheses that experimentalists can test, such as looking at the impact of lipids on cancer or finding new diagnostic tools.
“We are able to break down the lipid types that are important or unimportant, which is a big reason why experiments in the past had conflicting results,” Lightstone said. “This model creates new things that we can look at and try to understand cancer, which is very complex and no longer believed to be a singular disease, but a collection of diseases.”
The data generated by the simulations resulted in findings, predictions and hypotheses that were tested and validated via experiments at FNLCR. Cancer researchers are determining the compositions are making a difference.
“The simulations generated insights into the molecular details of the process by which KRAS promotes cancer,” said Director of FNLCR’s Cancer Research Technology Program Dwight Nissley, NCI’s lead for the JDACS4C Pilot 2 project. “Further studies will focus on mechanisms of cancer initiation that may reveal new therapeutic opportunities.”
Knowledge gained from the experiments will feed back into the machine learning-based MuMMI model, creating a validation loop that will make it more accurate, researchers said.
The work has continued with two more campaigns, adding RAF proteins, different variants of RAS, and computational advancements, including a new grand canonical version of the macro model, a new machine learning algorithm that can handle different cases and an additional third all-atom model scale. The latter development is the subject of future publications, including a recent paper describing the updated workflow, which was published by the 2021 International Conference for High Performance Computing, Networking, Storage and Analysis (SC21).
Researchers said the MuMMI framework could also be used for other simulation systems and have made the methodology available as open-source software on Github for other groups to develop their own multi-scaling methods.