An emergent role for radiomic decision support in lung cancer
Images
SA-CME credits are available for this article here.
Medical images represent anatomical and/or functional facsimiles of the human body. As such, they serve a critical role in the diagnosis of diseases and the evaluation of treatment response. Current interpretations of the images by radiologists comprise an anthropogenic synopsis of 2-dimensional (2D) or 3-dimensional (3D) spatial data. Despite extensive efforts at standardization, evaluations continue to depend on the individual evaluating the images, resulting in variation of interpretation.
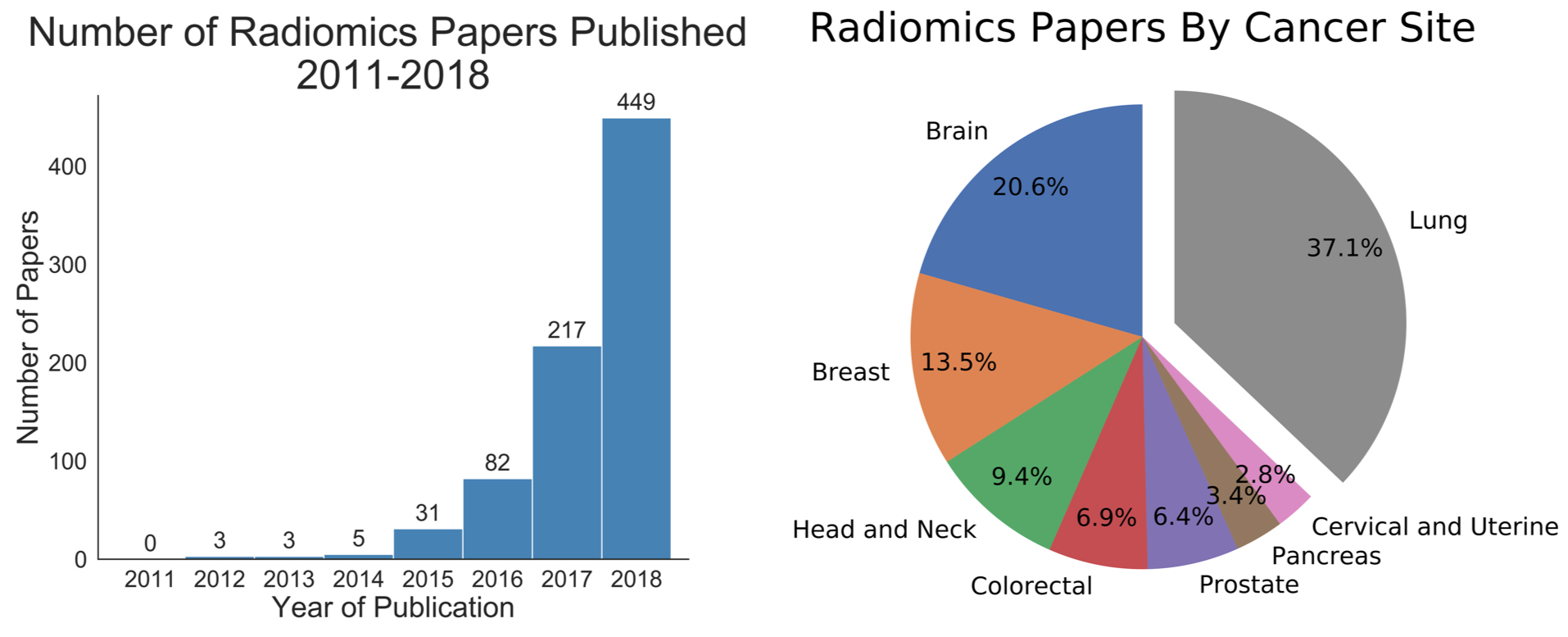
Radiomics is an emergent methodology within image analysis in which quantitative data is acquired using automated analysis techniques (Figure 1).1-4 The extracted information, also known as image features, can be combined with orthogonal data (eg, clinical data or biological measures [ie, mutations, transcriptomic panels, etc.]) to build prediction models for diagnosis or treatment selection. These strategies are poised to offer a more quantitative and objective basis for informed medical decision-making.1,5,6
The tripartite mainstays of cancer treatment include radiation therapy, chemotherapy, and surgery. These treatments extensively utilize medical images for diagnosis and to monitor efficacy. The imaging modalities most commonly used include computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET). The frequent utilization of these technologies provides clinical practices, even those with modest patient volumes, an extensive collection of mineable image data. Indeed, radiomics features have already been associated with improved diagnosis accuracy in cancer,7 specific gene mutations,8 and treatment responses to chemotherapy and/or radiation therapy in the brain,9,10 head and neck,11,12 lung,13-17 breast,18,19 and abdomen.20 More recently, radiomics features integrated into a multitasked neural network were combined with clinical data to derive a personalized radiation dose for patients treated with stereotactic lung radiation therapy.21 Altogether, these developments suggest that the integration of image data to inform clinical care is on the horizon.
Herein, we review recent developments in radiomics, its applications to lung cancer treatments, and the challenges associated with radiomics as a tool for precision diagnostics and theranostics.
Methodology
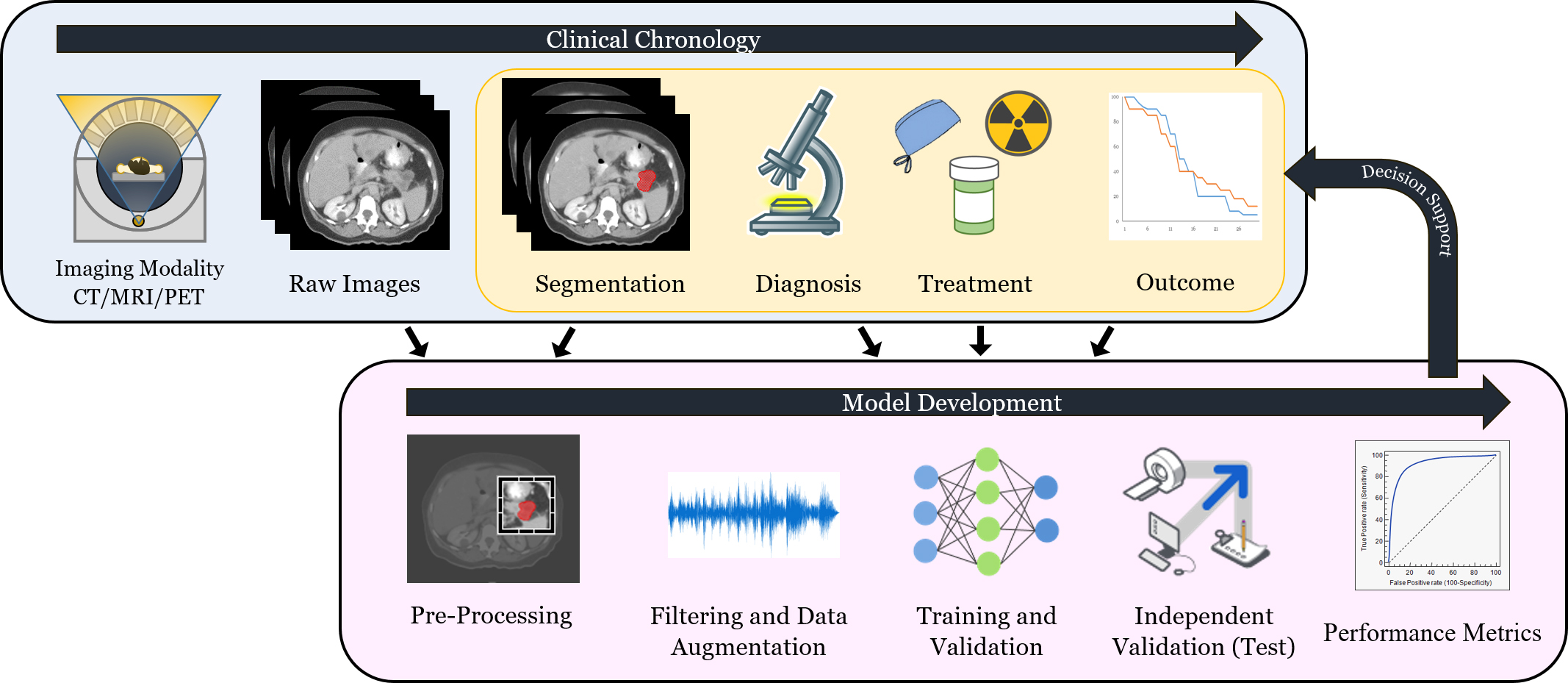
A general workflow of radiomics is depicted in Figure 2. At the data collection stage, imaging data is combined with clinical and histopathological data. Image data must undergo additional steps before downstream analyses, however, including region-of-interest segmentation, and feature and texture extraction. Based on the classification task at hand (eg, local failure after radiation, progression-free survival after immunotherapy, etc.), researchers can then proceed to the next stage, the training and validation of the radiomics model. After training and validation, a dataset that the algorithm has not yet seen (test or holdout set) is used to evaluate the model. If the model is shown to be accurate, it may potentially provide clinicians with improved decision-making capabilities. Transportability testing (using a dataset from a distinct but plausibly related population) of the model is also critical since it can help determine whether the model can be implemented more broadly in other settings. To establish transportability, an independent dataset external to the primary institution should be used.
Data Collection
The first step in radiomics is data acquisition. A large sample size is required because of the complexity of the prediction task. Since machine learning and neural network-based models can learn multifactorial, nonlinear relationships between image-based predictors and outcomes, models can inadvertently too closely fit or “memorize” the data they are built on. This can lead to poor performance on previously unseen data, a phenomenon known as overfitting. To mitigate overfitting, large datasets and other strategies are implemented to build improved and more generalizable models.
Although first developed using CT images, radiomic methodologies have also been implemented for other modalities such as MRI, PET, and ultrasound (US). Models are usually built on a single modality to ensure the consistent treatment of images in the preprocessing pipeline. Images and clinical data used to build a radiomic model can be gathered from single or multiple institutions. To ensure standardization among the data presented to the model, there are critical quality assurance steps at both the data acquisition and preprocessing steps. Standardization of imaging protocols and having a clearly defined, universally applicable preprocessing pipeline are critical for model reproducibility.
At the time of imaging, acquisition and reconstruction parameters such as voxel size and gray-level discretization are central to achieving reproducible results. Other factors that may affect stability of radiomic features include respiratory motion and use of IV contrast. It has been previously shown that inter-CT scanner variability22 and variability of random noise23 may affect the stability of radiomic features. To decrease variability of the features during the collection process, resampling and image cropping to a uniform spacing and size prior to extracting features is recommended.24-26 Another data optimization technique involves clipping and normalizing voxel intensities. Lastly, data augmentation through preprocessing transformations or data generation using neural networks can increase the data available to a nascent radiomic model.27
Segmentation
Delineation of the tumor and normal tissue is a crucial first step in both radiation therapy and radiomics, directly influencing the performance of radiomic models.28 Appropriate segmentation is critical to models that extract predefined features directly, as well as to neural models, which can be trained to emphasize the designated areas. Identifying the section of the image to be used for segmentation and extraction of radiomic features is a topic of ongoing investigation. Traditionally, features are extracted from the segmented tumor region. However, there is also increasing interest in image characteristics adjacent and external to the gross tumor volume. For example, Dou et al29 have shown the possibility to improve multivariate models to predict the risk of distant metastasis by extracting features from the peritumoral region.
There are certain obvious challenges with manual segmentation: Tumors may be near tissue with similar characteristics, making it difficult to distinguish between the two structures. Moreover, medical images may have distortions due to random noise, imaging resolution, and artifacts. To reduce intra- and interobserver variability, automatic or semi-automatic segmentation may improve the stability of radiomic features. Various methods have been proposed for semi-automatic segmentation.30,31 With recent advances in deep-learning algorithms, fully automatic segmentation methods have also been developed.32
Feature Extraction
Originally, radiomic models were developed from predefined, “handcrafted” features consisting of algebraic representations of voxel intensities. This structured data can be analyzed with classical statistics or with machine learning and neural networks. More recently, convolutional neural networks have been implemented to directly learn properties of the image, allowing for the extraction of features beyond those conceived and crafted by humans. Aspects from either or both methodologies can then be merged into a representative quantity (or quantities) known as an image signature.
Features can be categorized based on origin. Semantic features are those currently used in clinical practice as visualized and described by the radiologist. Radiomics complements these with nonsemantic, quantitatively and systematically extracted features, based on voxel intensity. Classic quantitative radiomic features can be further categorized as structural, first, second, and higher order. Structural features are the most basic descriptive and derived measures such as tumor volume, shape, maximum diameter, and surface area. These features can help quantify tumor spiculation and other factors that may indicate tumor malignancy. First-order features refer to simple statistical quantities such as mean, median, and maximum gray-level values found within the segmented tumor. Extracting second-order, or textural features, quantifies statistical inter-relationships between neighboring voxels. This provides a measure of spatial relationship between the voxel intensities in the tumor, which may allow for the determination of tissue heterogeneity.33 Higher-order statistical features are extracted by applying filters and transformations to the image. Two of the most popular methods are the Laplacian transforms of Gaussian-filtered images and wavelet transforms. Such higher-order methods increase the number of features extracted by the order of magnitude of filters applied. This allows identification of image attributes based on various spatial frequency patterns. Lastly, incorporating the change in radiomic features over time, or delta radiomic features, has been shown to improve lung cancer incidence,34 overall survival, and metastases prediction.35
Deep learning, a subset of machine learning, uses a neural network model that mimics the connectivity of a biological brain to identify complex abstractions of patterns using nonlinear transformations. Neural network models learn directly from unstructured data such as images through convolutional layers that synthesize voxel intensities into representative features. Deep-learning approaches typically require more data, a challenge that can be mitigated through various techniques such as data augmentation36 and transfer learning.37
Feature Selection
Manual feature extraction can result in thousands of radiomic features, some of which are redundant. In a dataset with clinical events (eg, local failure after radiation therapy) occurring at much lower magnitudes, inclusion of large-scale parameters with low event rates can contribute to model overtraining or overfitting. Utilization of feature selection techniques can help alleviate this potential pitfall.
Radiomic feature selection methods focus on stability of features, feature independence, and feature relevance. The stability of features may be analyzed with a test-retest dataset in which multiple images of the same modality are taken over a relatively short period to test whether such features are reproducible.38 Feature independence is assessed by statistical methods testing the correlation between the features themselves, such as principal component analysis (PCA). Feature selection based on relevance can be done with a univariate approach, testing whether each individual feature is correlated with the outcome being investigated, or a multivariate approach, which analyzes the combined predictive power of the features.
Parmar et al11 used clustering as a method to contend with the large number of quantitative features. The high-dimensional feature space was reduced into radiomic clusters, with clusters being predictive of patient survival, tumor stage and histology. Alternatively, neural networks have been shown to learn increasingly detailed geometries in each subsequent convolutional layer, and can be used to generate a set of highly descriptive image features.39
Development of Predictive Models
A predictive model is then constructed from the extracted relevant features creating a “radiomic signature.” Depending on the task at hand, various prediction models can be utilized (eg, classification and survivability models). Classification models categorize data into known categories (eg, tumor is benign or malignant). Survivability models require additional time-related information about the patients being treated, and aim to predict the time to failure or survival of patients undergoing a certain treatment. One approach to predict time-to-event clinical outcomes is by making the image signature equivalent to the logarithm of the hazard ratio in a Cox regression model.21,40 Other machine-learning methods can then be used with either manually extracted features or the outputs of neural network models to derive prediction scores.
Validation
To show that the radiomic model is generalizable, it must be validated. Model validation on an independently obtained external dataset is recommended. The model is usually analyzed using the receiver operating characteristic (ROC) curve with the area under the curve (AUC) being the commonly reported value in discrimination analysis. Model validation should be repeated on a target population prior to its deployment to ensure transportability.
Challenges and Opportunities
The rapid proliferation of radiomics applications has fueled optimism that medical images can be utilized to better guide clinicians in the recommendation of optimal treatment strategies. As with every technique and technology, however, certain challenges require attention to create and implement a robust radiomics model.
Data Sharing
Collecting and sharing data over multiple institutions or hospitals is a significant limitation to model development and testing. A single institution or hospital typically does not have enough events to establish and test a transportable radiomics model. To address this need, multiple data-sharing networks have been established to house shared data such as the Cancer Imaging Archive41 and the Quantitative Imaging Network.42 Contributions of well-annotated data to the shared datasets or collaborations between multiple institutes are critical for future model development and implementation.
Data Standardization
In multi-institutional radiomics studies, it is rare that all institutes share the same imaging acquisition settings such as imaging modality, protocol, or reconstruction algorithm. Additionally, image segmentation and interpretation of the data may be highly subjective and prone to human variations. While a highly standardized dataset will more likely guarantee a consistent model and reproducible predictions, this is an impractical expectation of a large dataset, especially in a multi-institutional setting. Data cleaning and preprocessing can mitigate these challenges through selection of similarly annotated images, image resampling, retrospective segmentation, and even translation of one modality to another.43 Additionally, the robustness of radiomic models built on multi-institutional datasets can be inherently higher since they are less prone to overfitting caused by a single institutional standard.
Model Evaluation
Although radiomic models may be highly performant on the data on which they are built, prediction results may be affected when implemented into real clinical settings due to model under-or overfitting. Therefore, it is crucial to use independent, external datasets to evaluate the predictive power of the established radiomic signature. Additionally, the radiomic model should be trained on new data as the standard of care continues to improve for it to adapt to new treatment protocols and prognosis, as well as to better quantify its accuracy. A reliable method to maintain an up-to-date radiomics model can be as critical as establishing the initial model. As data-sharing archives41,42 (noted above) become more prevalent, the need for large volumes of current, external images will be met. Since radiomic models can be deployed through online or locally hosted software, they are highly movable even if the independent data on which they are evaluated is not.
Model Interpretation
Since radiomics is a fairly new concept and model structures are inherently abstruse (representing a black box), questions and concerns are often raised toward the ultimate implementation of radiomic models. Physician skills and intuition are honed over years of training and experience. There is anticipated to be a gulf of trust between physicians’ “gestalt” and experience-driven approaches with the current difficult-to-interpret output of artificial intelligence systems. Efforts to improve the interpretability of predictive models include feature selection through bootstrapping44 as well as development of saliency maps highlighting the relative importance of voxels to the predicted outcome.21 The implication that radiomic models manifest underlying biology by being able to classify histological subtypes45,46 and gene mutations47,48 makes the association between genetics and radiomics an active area of research. This type of integrative analyses of known risk factors is needed to explain the meaning of radiomic features. Promoting enhanced interpretability of radiomic and neural-network-derived models will be a critical step to catalyze implementation as a decision-support tool.
Potential Applications
A growing number of studies show the value of radiomics as a tool to augment clinical decision-making, with significant progress in applying radiomics to lung cancer diagnosis, treatment, and risk evaluation.
Investigative Models
Aerts et al38 created a radiomic signature prognostic of overall survival in independent cohorts of patients based on intensity, shape, textural, and wavelet features. The features were selected based on stability using test-retest CT scans, independence, and univariate predictive capability of the features before constructing a multivariate model including the top feature from each of the four feature groups. Several radiomics studies have shown diagnostic potential in CT-based models to discriminate cancerous tumors from benign nodules.
A number of studies have also applied radiomics to predict histology based on pretreatment CT images45,46 and radiogenomics to identify the tumors’ underlying gene expression.47,48 Currently, histological classification and genetic subtyping depend on biopsies and re-biopsies. If radiomics methods achieve clinical levels of accuracy, it may allow patients to forego numerous invasive biopsies. For example, Wang et al47 showed that it is possible to create a deep neural network using CT images to provide an accurate method to establish epidermal growth factor receptor (EGFR) status in lung adenocarcinoma patients, potentially reducing the need for biopsy.
Another set of studies looked at the prognostic and predictive possibilities of using the radiomic approach—an important area in precision medicine because it informs the creation of an optimal treatment plan. Such studies predict probability of response to treatment,49 survival,50,51 and risk of metastases.29,52
Extending classification and survivability models to guide treatment, Lou et al21 developed an image-based, deep-learning framework for the individualizing of radiation therapy dose. First a risk score was identified by a deep neural network, Deep Profiler. This signature outperformed classical radiomic features in predicting treatment outcome. This framework also incorporates a model to project optimized radiation dose to minimize treatment failure probability.
Hosny et al36 trained deep neural networks to stratify patients into low- and high-mortality risk groups, and were also able to outperform models based on classical radiomic features as well as clinical parameters. The neural network predictions were largely stable when tested against imaging artifacts and test-retest scans. In addition, there was a suggestion that deep-learning extracted features may be associated with biological pathways including cell cycle, DNA transcription, and DNA replication.
Altogether, radiomics could potentially serve an important complementary role to other orthogonal data such as genetic and clinical information to improve assessment of clinical characteristics and molecular information.
Deployment
The models discussed have translation potential because they could be integrated into clinical practice upon additional and prospective validation. Imaging is a mainstay of clinical use, and software deployment of radiomic models are noninvasive and, if designed with user input, can be seamlessly integrated into daily workflow for the intended specialist (eg, radiologist or radiation oncologist).
There are a several avenues of implementation for software facilitating radiomic analyses into routine clinical practice. These include improved segmentation through semi-automatic or automatic contouring, which can be achieved by traditional image analysis techniques such as region-growing,30,31 convolutional techniques such as neural network-based segmentation,32 or “smart-contouring” techniques based on the regions of an image determined to be salient based on a deep-learning model.21 Another promising area for integration is risk-profiling. Modeling risk can be achieved through a software package paired with an institution’s existing imaging server. This should minimize significant disruption of the existing clinical workflow. As with any method of risk-profiling, predictive radiomic models could serve as an advisory decision-support tool in the hands of the radiologist and radiation oncologist. Specifically, radiomic models that both model and mitigate the risks are poised to alter the clinical paradigm(s). Adjusting treatment strategies through dose-specific21 or targeted agent-specific recommendations represent possible uses that could improve clinical outcomes in select patient populations. As with segmentation and risk-profiling, these applications can be achieved through software deployment.
Lastly, while other biomarkers are likely to represent critical orthogonal inputs to more accurately predict clinical outcomes, it is possible that tumor intrinsic determinants (ie, genetic alterations, RNA gene expression, etc.) can be detected by radiomic features, as suggested.38,53 Additional studies that seek to determine whether these classes of variables (image vs biology) are tautological, orthogonal or somewhere in between will be critical to assessing the need for additional inputs into the models. Convergence toward an integrative approach that incorporates these varied inputs is likely unavoidable in order to improve model accuracy and ultimate clinical deployment.
Conclusions
Radiomics is a computational image evaluation technique that integrates medical images, clinical data, and machine learning. Despite hurdles to implementation, radiomic models show immense potential for personalized lung cancer diagnosis, risk profiling, and treatment due to their ability to incorporate image characteristics beyond the ken of the human observer.
References
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441-446. doi:10.1016/j.ejca.2011.11.036
- Parekh V, Jacobs MA. Radiomics: a new application from established techniques. Expert Rev Precis Med Drug Dev. 2016;1(2):207-226. doi:10.1080/23808993.2016.1164013
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563-577. doi:10.1148/radiol.20151511694.
- Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2(1). doi:10.1186/s41747-018-0068-z
- Bi WL, Hosny A, Schabath MB, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Cancer J Clin. February 2019. doi:10.3322/caac.21552
- Morin O, Vallières M, Jochems A, et al. A deep look into the future of quantitative imaging in oncology: a statement of working principles and proposal for change. Int J Radiat Oncol. 2018;102(4):1074-1082. doi:10.1016/j.ijrobp.2018.08.032
- Ardila D, Kiraly AP, Bharadwaj S, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med. 2019;25(6):954. doi:10.1038/s41591-019-0447-x
- Karlo CA, Di Paolo PL, Chaim J, et al. Radiogenomics of clear cell renal cell carcinoma: associations between ct imaging features and mutations. Radiology. 2014;270(2):464-471. doi:10.1148/radiol.13130663
- Zhang Z, Yang J, Ho A, et al. A predictive model for distinguishing radiation necrosis from tumour progression after Gamma Knife radiosurgery based on radiomic features from MR images. Eur Radiol. 2018;28(6):2255-2263. doi:10.1007/s00330-017-5154-8
- Kickingereder P, Burth S, Wick A, et al. Radiomic profiling of glioblastoma: identifying an imaging predictor of patient survival with improved performance over established clinical and radiologic risk models. Radiology. 2016;280(3):880-889. doi:10.1148/radiol.2016160845
- Parmar C, Leijenaar RTH, Grossmann P, et al. Radiomic feature clusters and prognostic signatures specific for lung and head & neck cancer. Sci Rep. 2015;5(1). doi:10.1038/srep11044
- Giraud P, Giraud P, Gasnier A, et al. Radiomics and machine learning for radiotherapy in head and neck cancers. Front Oncol. 2019;9. doi:10.3389/fonc.2019.00174
- Causey JL, Zhang J, Ma S, et al. Highly accurate model for prediction of lung nodule malignancy with CT scans. Sci Rep. 2018;8(1). doi:10.1038/s41598-018-27569-w
- Choi W, Oh JH, Riyahi S, et al. Radiomics analysis of pulmonary nodules in low-dose CT for early detection of lung cancer. Med Phys. 2018;45(4):1537-1549. doi:10.1002/mp.12820
- Chen L, Huang B, Huang X, Cao W, Sun W, Deng X. Clinical evaluation for the difference of absorbed doses calculated to medium and calculated to water by Monte Carlo method. Radiat Oncol. 2018;13(1). doi:10.1186/s13014-018-1081-3
- Han F, Wang H, Zhang G, et al. Texture feature analysis for computer-aided diagnosis on pulmonary nodules. J Digit Imaging. 2015;28(1):99-115. doi:10.1007/s10278-014-9718-8
- Liu K, Kang G. Multiview convolutional neural networks for lung nodule classification. Int J Imaging Syst Technol. 2017;27(1):12-22. doi:10.1002/ima.22206
- Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys. 2017;44(10):5162-5171. doi:10.1002/mp.12453
- Parekh VS, Jacobs MA. Integrated radiomic framework for breast cancer and tumor biology using advanced machine learning and multiparametric MRI. Npj Breast Cancer. 2017;3(1). doi:10.1038/s41523-017-0045-3
- Hou Z, Yang Y, Li S, et al. Radiomic analysis using contrast-enhanced CT: predict treatment response to pulsed low dose rate radiotherapy in gastric carcinoma with abdominal cavity metastasis. Quant Imaging Med Surg. 2018;8(4):410-420. doi:10.21037/qims.2018.05.01
- Lou B, Doken S, Zhuang T, et al. An image-based deep learning framework for individualising radiotherapy dose: a retrospective analysis of outcome prediction. Lancet Digit Health. 2019;1(3):e136-e147. doi:10.1016/S2589-7500(19)30058-5
- Mackin D, Fave X, Zhang L, et al. Measuring computed tomography scanner variability of radiomics features. Invest Radiol. 2015;50(11):757-765. doi:10.1097/RLI.0000000000000180
- Midya A, Chakraborty J, Gönen M, Do RKG, Simpson AL. Influence of CT acquisition and reconstruction parameters on radiomic feature reproducibility. J Med Imaging. 2018;5(01):1. doi:10.1117/1.JMI.5.1.011020
- Larue RTHM, van Timmeren JE, de Jong EEC, et al. Influence of gray level discretization on radiomic feature stability for different CT scanners, tube currents and slice thicknesses: a comprehensive phantom study. Acta Oncol. 2017;56(11):1544-1553. doi:10.1080/0284186X.2017.1351624
- Shafiq-ul-Hassan M, Latifi K, Zhang G, Ullah G, Gillies R, Moros E. Voxel size and gray level normalization of CT radiomic features in lung cancer. Sci Rep. 2018;8(1). doi:10.1038/s41598-018-28895-9
- Shafiq-ul-Hassan M, Zhang GG, Latifi K, et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med Phys. 2017;44(3):1050-1062. doi:10.1002/mp.12123
- Wang J, Mall S, Perez L. The effectiveness of data augmentation in image classification using deep learning. In: Standford University research report, 2017.
- Haga A, Takahashi W, Aoki S, et al. Classification of early stage non-small cell lung cancers on computed tomographic images into histological types using radiomic features: interobserver delineation variability analysis. Radiol Phys Technol. 2018;11(1):27-35. doi:10.1007/s12194-017-0433-2
- Dou TH, Coroller TP, van Griethuysen JJM, Mak RH, Aerts HJWL. Peritumoral radiomics features predict distant metastasis in locally advanced NSCLC. Lee H-S, ed. PLOS ONE. 2018;13(11):e0206108. doi:10.1371/journal.pone.0206108
- Lassen BC, Jacobs C, Kuhnigk J-M, Ginneken B van, Rikxoort EM van. Robust semi-automatic segmentation of pulmonary subsolid nodules in chest computed tomography scans. Phys Med Biol. 2015;60(3):1307-1323. doi:10.1088/0031-9155/60/3/1307
- Velazquez ER, Parmar C, Jermoumi M, et al. Volumetric CT-based segmentation of NSCLC using 3D-slicer. Sci Rep. 2013;3(1). doi:10.1038/srep03529
- Yang X, Pan X, Liu H, et al. A new approach to predict lymph node metastasis in solid lung adenocarcinoma: a radiomics nomogram. J Thorac Dis. 2018;10(S7):S807-S819. doi:10.21037/jtd.2018.03.126
- Lubner MG, Smith AD, Sandrasegaran K, Sahani DV, Pickhardt PJ. CT texture analysis: definitions, applications, biologic correlates, and challenges. RadioGraphics. 2017;37(5):1483-1503. doi:10.1148/rg.2017170056
- Cherezov D, Hawkins SH, Goldgof DB, et al. Delta radiomic features improve prediction for lung cancer incidence: a nested case-control analysis of the National Lung Screening Trial. Cancer Med. 2018;7(12):6340-6356. doi:10.1002/cam4.1852
- Fave X, Zhang L, Yang J, et al. Delta-radiomics features for the prediction of patient outcomes in non–small cell lung cancer. Sci Rep. 2017;7(1). doi:10.1038/s41598-017-00665-z
- Hosny A, Parmar C, Coroller TP, et al. Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. Butte AJ, ed. PLOS Med. 2018;15(11):e1002711. doi:10.1371/journal.pmed.1002711
- Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345-1359. doi:10.1109/TKDE.2009.191
- Aerts HJWL, Velazquez ER, Leijenaar RTH, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5(1). doi:10.1038/ncomms5006
- Zeiler MD, Fergus R. Visualizing and understanding convolutional networks. ArXiv13112901 Cs. November 2013. http://arxiv.org/abs/1311.2901. Accessed August 1, 2019.
- Katzman JL, Shaham U, Cloninger A, Bates J, Jiang T, Kluger Y. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med Res Methodol. 2018;18(1). doi:10.1186/s12874-018-0482-1
- Prior F, Smith K, Sharma A, et al. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci Data. 2017;4(1). doi:10.1038/sdata.2017.124
- Clarke LP, Nordstrom RJ, Zhang H, et al. The Quantitative Imaging Network: NCI’s historical perspective and planned goals. Transl Oncol. 2014;7(1):1-4. doi:10.1593/tlo.13832
- Lei Y, Harms J, Wang T, et al. MRI-only based synthetic CT generation using dense cycle consistent generative adversarial networks. Med Phys. 2019;46(8):3565-3581. doi:10.1002/mp.13617
- Peikert T, Duan F, Rajagopalan S, et al. Novel high-resolution computed tomography-based radiomic classifier for screen-identified pulmonary nodules in the National Lung Screening Trial. PLOS ONE. 2018;13(5):e0196910. doi:10.1371/journal.pone.0196910
- Wu W, Parmar C, Grossmann P, et al. Exploratory Study to identify radiomics classifiers for lung cancer histology. Front Oncol. 2016;6. doi:10.3389/fonc.2016.00071
- Ferreira Junior JR, Koenigkam-Santos M, Cipriano FEG, Fabro AT, Azevedo-Marques PM de. Radiomics-based features for pattern recognition of lung cancer histopathology and metastases. Comput Methods Programs Biomed. 2018;159:23-30. doi:10.1016/j.cmpb.2018.02.015
- Wang S, Shi J, Ye Z, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53(3):1800986.doi:10.1183/13993003.00986-2018
- Liu Y, Kim J, Balagurunathan Y, et al. Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer. 2016;17(5):441-448.e6. doi:10.1016/j.cllc.2016.02.001
- Aerts HJWL, Grossmann P, Tan Y, et al. Defining a radiomic response phenotype: a pilot study using targeted therapy in NSCLC. Sci Rep. 2016;6(1). doi:10.1038/srep33860
- Huynh E, Coroller TP, Narayan V, et al. CT-based radiomic analysis of stereotactic body radiation therapy patients with lung cancer. Radiother Oncol. 2016;120(2):258-266. doi:10.1016/j.radonc.2016.05.024
- Huang Y, Liu Z, He L, et al. Radiomics signature: a potential biomarker for the prediction of disease-free survival in early-stage (i or ii) non-small cell lung cancer. Radiology. 2016;281(3):947-957. doi:10.1148/radiol.2016152234
- Coroller TP, Grossmann P, Hou Y, et al. CT- based radiomic signature predicts distant metastasis in lung adenocarcinoma. Radiother Oncol. 2015;114(3):345-350. doi:10.1016/j.radonc.2015.02.015
- Bakr S, Gevaert O, Echegaray S, et al. A radiogenomic dataset of non-small cell lung cancer. Sci Data. 2018;5:180202. doi:10.1038/sdata.2018.202
Citation
GA K, M G, T M, T Z, ME A. An emergent role for radiomic decision support in lung cancer. Appl Radiat Oncol. 2019;(4):24-30.
December 28, 2019